SEGPROJ¶
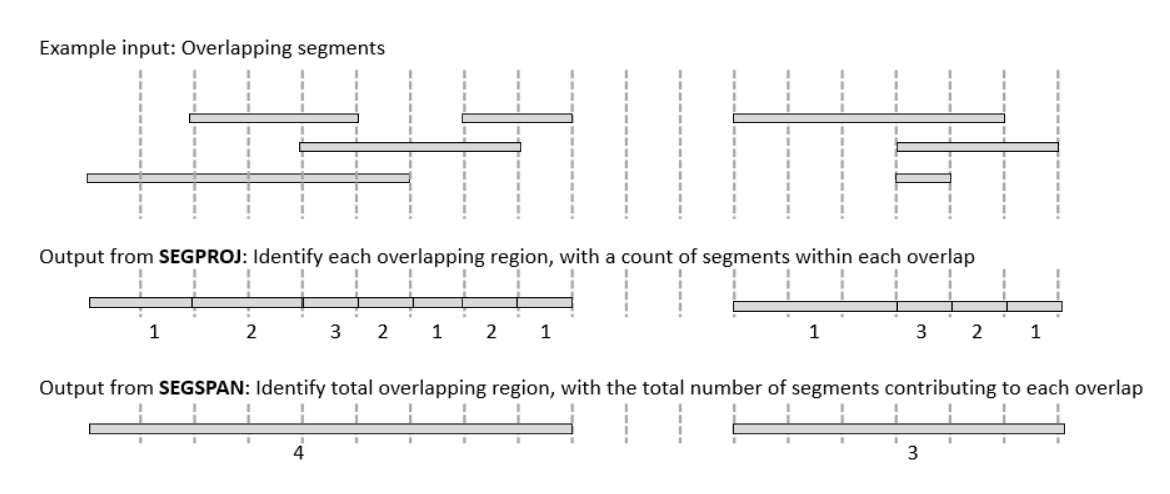
The SEGPROJ command takes in a stream of segments, e.g. (chrom,bpstart,bpstop), and projects them. It returns a stream of segments, with additional column segCount, where each segments represents a region with a constant count of overlapping segments.
Overlapping Variants module in the Sequence Miner¶
Usage¶
gor ... | SEGPROJ [-maxseg bpsize] [-f bpsize] [-gc cols]
Options¶
|
The maximum span of the segments in the input stream. Defaults to 4000000 (4Mb). |
|
Fuzzfactor to expand the input segments (upstream and downstream). |
|
Grouping columns, i.e. only segments within each group are projected together. |
|
Summing column. Specify a column with integer values used for the sum operation. |
Examples¶
Using SEGPROJ on a simple overlap of regions.
- Input:
Chrom, start, stop chr1, 100, 1000 chr1, 500, 1200
gor ... | segproj
- Output:
Chrom, start, stop chr1, 100, 500, 1 chr1, 500, 1000, 2 chr1, 1000, 1200, 1
- Input:
Chrom, start, stop chr1, 100, 1000, 1 chr1, 500, 1200, 2
gor ... | segproj -sumcol 4
- Output:
Chrom, start, stop chr1, 100, 500, 1 chr1, 500, 1000, 3 (1+2) chr1, 1000, 1200, 2
gor ref/ensgenes/ensexons.gorz | segproj -f 10 -maxseg 100000 | where segCount > 1
The above query finds regions where two or more exons overlap with the proximity of 10 bases.
gor ref/ensgenes/ensgenes.gorz | segproj -gc strand -maxseg 3000000
The above query returns segments showing the overlap genes on the same strand.
gor #CNVs# -f PN1,PN2,PN3 | segspan -gc PN -maxseg 1000000 | segproj -maxseg 1000000 | join -segseg -maxseg 1000000 <(gor #CNVs# -f PN1,PN2,PN3 | segspan -gc PN -maxseg -maxseg 1000000) | group 1 -gc bpstop -lis -sc PN
The above query shows regions of overlap between CNVs from three subjects and lists the PN ids of the subjects having CNVs in the corresponding region.