RANK¶
The RANK command returns the rank of a number within a set of numbers. It compares the value of each cell in the specified column to all the values in the result set and returns the rank of that row in a column labelled “rank_<column_name>”.
The RANK command has two necessary parameters, namely the binsize, which can be set to “chrom”, “genome” or to a numeric value. The column specified in the RANK command must be a numeric column.
Using RANK in a nor query will give an error if binsize input parameter is used.
Options¶
|
Rank order. By default it is descending. |
|
Report rank distribution, lower rank and equal. |
|
Report z-value = (x-mean)/std. |
|
Report the total count for the bin. |
|
Report the value where the rank is 1. |
|
Report only rows where rank <= number. |
|
Grouping columns (other than bin). |
Examples¶
The example below takes some entries from the #dbsnp# table with indels of some length, calculates the length of the reference column in the row and then ranks the value in the column.
gor #dbsnp# | WHERE len(reference) > 4 OR len(allele) > 4
| CALC refLength len(reference) | PREFIX refLength calc | TOP 6
| RANK genome calc_refLength
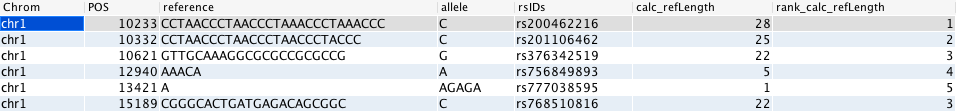
The following query will perform a parallelised gor query that calculates the length of each gene and then ranks them by length and returns the longest gene on each chromosome.
pgor #genes#
| calc length (gene_end - gene_start)
| rank chrom length -o desc
| where rank_length = 1
The next query will return the shortest gene:
pgor #genes#
| calc length (gene_end - gene_start)
| rank chrom length -o asc
| where rank_length = 1